Photo Credits – www.genome.gov
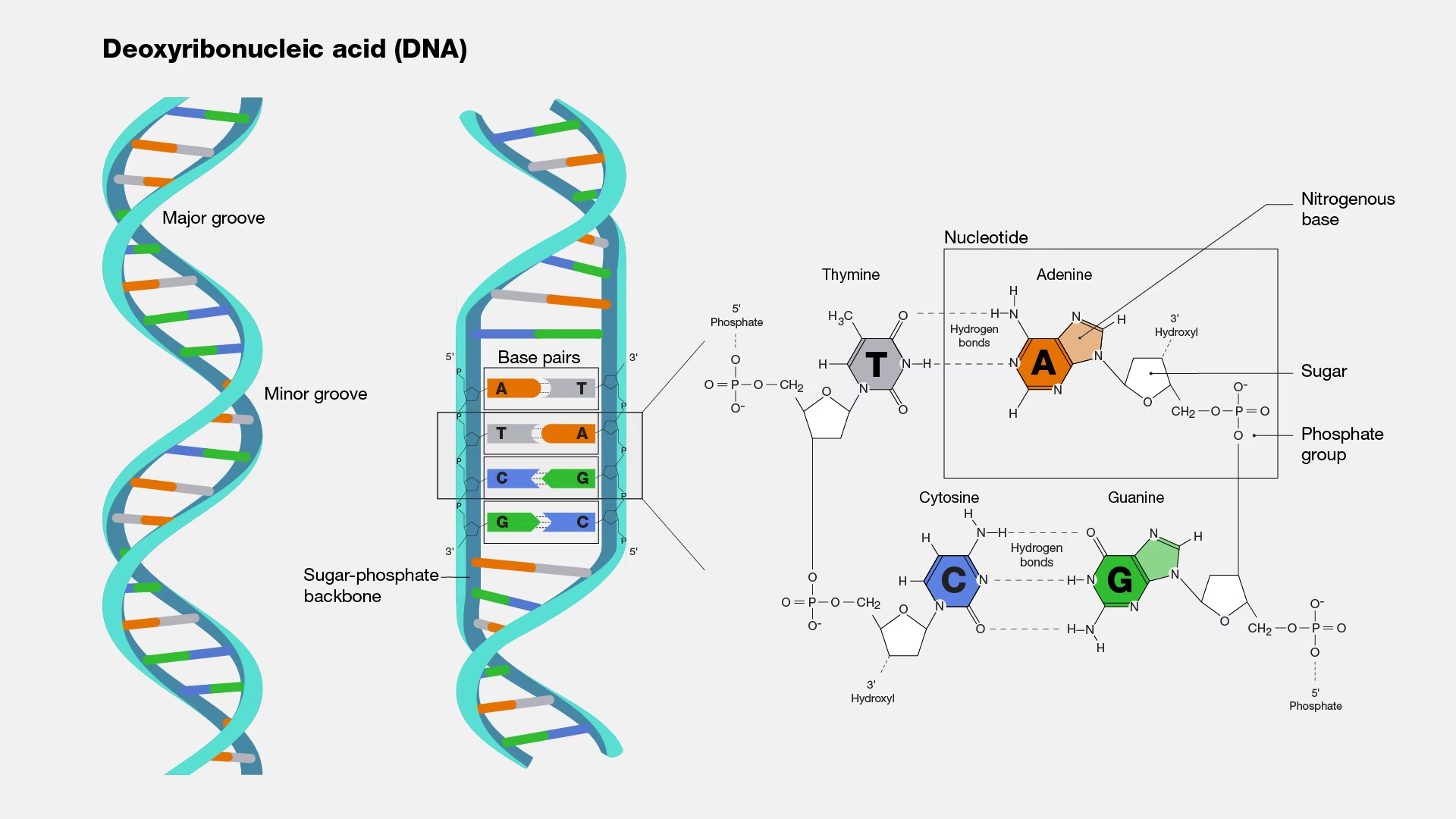
DNA, or deoxyribonucleic acid, is the molecule that carries genetic information essential for an organism’s development and function. It consists of two strands forming a double helix, with a backbone of sugar (deoxyribose) and phosphate groups. Four bases i.e. adenine (A), cytosine (C), guanine (G), and thymine (T), attach to the sugars, pairing specifically (A with T, and C with G) to connect the strands. The sequence of these bases encodes biological instructions for producing proteins or RNA.
What does half-life of DNA means?
The half-life of DNA refers to the time it takes for half of the molecular bonds in a DNA strand to break down under specific conditions. This process is influenced by factors such as temperature, moisture, and the surrounding environment. In optimal conditions, such as permafrost or dry caves, fragments of DNA can endure for tens of thousands of years. However, In a harsher environment it may degrade within mere decades.
Understanding DNA’s half-life is more than a scientific curiosity. From reconstructing ancient genomes and tracing evolutionary pathways to solving historical mysteries, the ability to measure and predict DNA’s decay timeline has revolutionized fields like archaeology, paleogenetics, and forensic science.

Study – Half-Life of DNA in bone decay
A study on bone decay done in 2012, analysed mitochondrial DNA (mtDNA) from 158 radiocarbon-dated moa bones to confirm an exponential decay model of DNA degradation. It estimated a DNA half-life of 521 years for a 242 bp mtDNA sequence at 13.1°C, with a fragmentation rate much slower than in vitro predictions. While the exponential model fits well, DNA preservation varied significantly between samples, influenced by factors like taphonomy and bone diagenesis rather than age. Nuclear DNA was found to degrade at least twice as fast as mtDNA. These findings offer a baseline for predicting DNA survival in fossils.
Genes are like the story, and DNA is the language that the story is written in.
Now, the question would in your mind, that if the DNA has a half-life of 521 years, how are we able to accurately sequence the genome of, say, Neanderthals, who died out tens of thousands of years ago? How is DNA preserved in the bone marrow for such a long time?
Understanding the calculation
Let’s start with our answer.
Well, it does seem a little crazy that after say 40,000 years (about when Neanderthals were alive) any DNA from any cell should survive. However, we have numbers on our side. There are about 3.2 billion (3.2×109) base pairs in the human genome, and bone marrow is packed full of cells (more than a million per ml). Each of those cells has its diploid complement of chromosomes, bringing the total up to around 6.4×109 bp each. So even though after all that time (80 halvings) the contribution of any one cell is going to be minimal, there will be (hopefully) enough fragments of DNA contributed by multiple cells to stitch them all together to cover the whole genome.
DNA is a pretty stable molecule, provided it’s kept the right way, and it just so happens that the inside of a (dry or frozen) bone can sometimes provide those conditions. I’ve also seen talks from researchers who use the blood inside the root of a tooth, as the enamel casing acts as a pretty decent shield.
let us deep dive even further;
Half-life works by the length of time for half of the original sample to decay. So, for example, how long it would take 1 gram of radioactive substance to decay to 1/2 gram radioactive and 1/2 gram of a more stable version. So, for DNA, after 521 years half of the original DNA has decayed, but the other half is still there.
With a half-life of 521 years, and Neanderthal going extinct about 40,000 years ago…then that comes out to approximately 77 half-life periods.
40,000/521=76.77.
So, with each period half of the DNA’s nucleotide bonds have broken. After 77 periods that means that there would be:
Half-life periods of n=77, so the percent of the original sample remaining equals to 100/2^n.
100/(2^77) = 6.617*10^-22%
So, there would be 6.6×10-22 % left of the original sample.
So, not the same part of the DNA’S from each cell decays at the same rate. There might be an extremely small fraction remaining, but that fraction is different from different cells. Therefore, we can use these various remaining pieces to sequence the genome.





0 Comments